java-libsvm
Package provides the direct java conversion of the origin libsvm
Install
Add the following dependency to your POM file:
<dependency>
<groupId>com.github.chen0040</groupId>
<artifactId>java-libsvm</artifactId>
<version>1.0.4</version>
</dependency>
Usage
The package use data frame as containers for training and testing data (Please refers to this link on how to create a data frame from file or from scratch)
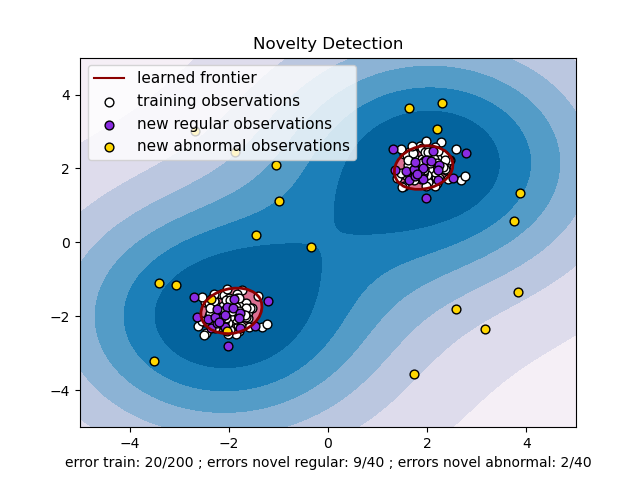
One-class SVM
Below is the code to create and train a one-class SVM:
OneClassSVM algorithm = new OneClassSVM();
algorithm.fit(training_data)
Below is the code to predict if data point is an outlier:
algorithm.isAnomaly(data_point)
SVR
Below is the code to create and train a SVR for regression modelling:
SVR algorithm = new SVR();
algorithm.fit(training_data)
Below is the code to perform data regression prediction:
algorithm.isAnomaly(data_point)
BinarySVC
Below is the code to create and train a BinarySVC for binary classification:
BinarySVC algorithm = new BinarySVC();
algorithm.fit(training_data)
Below is the code to perform data binary classification:
algorithm.isInClass(data_point)
OneVsOneSVC
Below is the code to create and train a OneVsOneSVC for multi-class classification:
BinarySVC algorithm = new BinarySVC();
algorithm.fit(training_data)
Below is the code to perform multi-class classification:
algorithm.classify(data_point)
Data Format
The data format default is the DataFrame class, which can be used to load csv and libsvm format text file. Please refers to the unit test cases on how they can be used.
Sample codes
Sample code for OneClassSVM:
Below is a sample code example of the one-class SVM for the example below here:
import com.github.chen0040.data.frame.DataFrame;
import com.github.chen0040.data.frame.DataQuery;
import com.github.chen0040.data.frame.Sampler;
import com.github.chen0040.svmext.oneclass.OneClassSVM;
DataQuery.DataFrameQueryBuilder schema = DataQuery.blank()
.newInput("c1")
.newInput("c2")
.newOutput("anomaly")
.end();
Sampler.DataSampleBuilder negativeSampler = new Sampler()
.forColumn("c1").generate((name, index) -> randn() * 0.3 + (index % 2 == 0 ? -2 : 2))
.forColumn("c2").generate((name, index) -> randn() * 0.3 + (index % 2 == 0 ? -2 : 2))
.forColumn("anomaly").generate((name, index) -> 0.0)
.end();
Sampler.DataSampleBuilder positiveSampler = new Sampler()
.forColumn("c1").generate((name, index) -> rand(-4, 4))
.forColumn("c2").generate((name, index) -> rand(-4, 4))
.forColumn("anomaly").generate((name, index) -> 1.0)
.end();
DataFrame trainingData = schema.build();
trainingData = negativeSampler.sample(trainingData, 200);
System.out.println(trainingData.head(10));
DataFrame crossValidationData = schema.build();
crossValidationData = negativeSampler.sample(crossValidationData, 40);
DataFrame outliers = schema.build();
outliers = positiveSampler.sample(outliers, 40);
final double threshold = 0.5;
OneClassSVM algorithm = new OneClassSVM();
algorithm.set_gamma(0.1);
algorithm.set_nu(0.1);
algorithm.thresholdSupplier = () -> 0.0;
algorithm.fit(trainingData);
for(int i = 0; i < crossValidationData.rowCount(); ++i){
boolean predicted = algorithm.isAnomaly(crossValidationData.row(i));
logger.info("predicted: {}\texpected: {}", predicted, crossValidationData.row(i).target() > threshold);
}
for(int i = 0; i < outliers.rowCount(); ++i){
boolean predicted = algorithm.isAnomaly(outliers.row(i));
logger.info("outlier predicted: {}\texpected: {}", predicted, outliers.row(i).target() > threshold);
}
Sample codes for SVR
Below is another complete sample code of the SVR to predict y = 4 + 0.5 * x1 + 0.2 * x2:
import com.github.chen0040.data.frame.DataFrame;
import com.github.chen0040.data.frame.DataQuery;
import com.github.chen0040.data.frame.Sampler;
import com.github.chen0040.svmext.oneclass.SVR;
DataQuery.DataFrameQueryBuilder schema = DataQuery.blank()
.newInput("x1")
.newInput("x2")
.newOutput("y")
.end();
// y = 4 + 0.5 * x1 + 0.2 * x2
Sampler.DataSampleBuilder sampler = new Sampler()
.forColumn("x1").generate((name, index) -> randn() * 0.3 + index)
.forColumn("x2").generate((name, index) -> randn() * 0.3 + index * index)
.forColumn("y").generate((name, index) -> 4 + 0.5 * index + 0.2 * index * index + randn() * 0.3)
.end();
DataFrame trainingData = schema.build();
trainingData = sampler.sample(trainingData, 200);
System.out.println(trainingData.head(10));
DataFrame crossValidationData = schema.build();
crossValidationData = sampler.sample(crossValidationData, 40);
SVR svr = new SVR();
svr.fit(trainingData);
for(int i = 0; i < crossValidationData.rowCount(); ++i){
double predicted = svr.transform(crossValidationData.row(i));
double actual = crossValidationData.row(i).target();
System.out.println("predicted: " + predicted + "\texpected: " + actual);
}
Sample code for BinarySVC
Below is another complete sample code of the BinarySVC for binary classification:
import com.github.chen0040.data.frame.DataFrame;
import com.github.chen0040.data.frame.DataQuery;
import com.github.chen0040.data.frame.Sampler;
import com.github.chen0040.svmext.classifiers.BinarySVC;
DataQuery.DataFrameQueryBuilder schema = DataQuery.blank()
.newInput("c1")
.newInput("c2")
.newOutput("anomaly")
.end();
Sampler.DataSampleBuilder negativeSampler = new Sampler()
.forColumn("c1").generate((name, index) -> randn() * 0.3 + (index % 2 == 0 ? -2 : 2))
.forColumn("c2").generate((name, index) -> randn() * 0.3 + (index % 2 == 0 ? -2 : 2))
.forColumn("anomaly").generate((name, index) -> 0.0)
.end();
Sampler.DataSampleBuilder positiveSampler = new Sampler()
.forColumn("c1").generate((name, index) -> rand(-4, 4))
.forColumn("c2").generate((name, index) -> rand(-4, 4))
.forColumn("anomaly").generate((name, index) -> 1.0)
.end();
DataFrame trainingData = schema.build();
trainingData = negativeSampler.sample(trainingData, 200);
trainingData = positiveSampler.sample(trainingData, 200);
System.out.println(trainingData.head(10));
DataFrame crossValidationData = schema.build();
crossValidationData = negativeSampler.sample(crossValidationData, 40);
crossValidationData = positiveSampler.sample(crossValidationData, 40);
BinarySVC algorithm = new BinarySVC();
algorithm.fit(trainingData);
BinaryClassifierEvaluator evaluator = new BinaryClassifierEvaluator();
for(int i = 0; i < crossValidationData.rowCount(); ++i){
boolean predicted = algorithm.isInClass(crossValidationData.row(i));
boolean actual = crossValidationData.row(i).target() > 0.5;
evaluator.evaluate(actual, predicted);
System.out.println("predicted: " + predicted + "\texpected: " + actual);
}
evaluator.report();
Sample codes for OneVsOneSVC
Below is another complete sample code of the OneVsOneSVC for multi-class classification:
import com.github.chen0040.data.frame.DataFrame;
import com.github.chen0040.data.frame.DataQuery;
import com.github.chen0040.data.frame.Sampler;
import com.github.chen0040.svmext.classifiers.OneVsOneSVC;
InputStream irisStream = new FileInputStream("iris.data");
DataFrame irisData = DataQuery.csv(",", false)
.from(irisStream)
.selectColumn(0).asNumeric().asInput("Sepal Length")
.selectColumn(1).asNumeric().asInput("Sepal Width")
.selectColumn(2).asNumeric().asInput("Petal Length")
.selectColumn(3).asNumeric().asInput("Petal Width")
.selectColumn(4).asCategory().asOutput("Iris Type")
.build();
TupleTwo<DataFrame, DataFrame> parts = irisData.shuffle().split(0.9);
DataFrame trainingData = parts._1();
DataFrame crossValidationData = parts._2();
System.out.println(crossValidationData.head(10));
OneVsOneSVC multiClassClassifier = new OneVsOneSVC();
multiClassClassifier.fit(trainingData);
ClassifierEvaluator evaluator = new ClassifierEvaluator();
for(int i=0; i < crossValidationData.rowCount(); ++i) {
String predicted = multiClassClassifier.classify(crossValidationData.row(i));
String actual = crossValidationData.row(i).categoricalTarget();
System.out.println("predicted: " + predicted + "\tactual: " + actual);
evaluator.evaluate(actual, predicted);
}
evaluator.report();